Get ahead
VMware offers training and certification to turbo-charge your progress.
Learn moreIn the first part of this blog series, we explored the basics of integrating Spring AI with large language models. We walked through building a custom ChatClient, leveraging Function Calling for dynamic interactions, and refining our prompts to suit the Spring Petclinic use case. By the end, we had a functional AI assistant capable of understanding and processing requests related to our veterinary clinic domain.
Now, in Part II, we’ll go a step further by exploring Retrieval-Augmented Generation (RAG), a technique that enables us to handle large datasets that wouldn’t fit within the constraints of a typical Function Calling approach. Let’s see how RAG can seamlessly integrate AI with domain-specific knowledge.
While listing veterinarians could have been a straightforward implementation, I chose this as an opportunity to showcase the power of Retrieval-Augmented Generation (RAG).
RAG integrates large language models with real-time data retrieval to produce more accurate and contextually relevant text. Although this concept aligns with our previous work, RAG typically emphasizes data retrieval from a vector store.
A vector store contains data in the form of embeddings—numerical representations that capture the meaning of the information, such as the data about our veterinarians. These embeddings are stored as high-dimensional vectors, facilitating efficient similarity searches based on semantics rather than traditional text-based searches.
For instance, consider the following veterinarians and their specialties:
Dr. Alice Brown - Cardiology
Dr. Bob Smith - Dentistry
Dr. Carol White - Dermatology
In a conventional search, a query for "Teeth Cleaning" would yield no exact matches. However, with semantic search powered by embeddings, the system recognizes that "Teeth Cleaning" relates to "Dentistry." Consequently, Dr. Bob Smith would be returned as the best match, even though his specialty was never explicitly mentioned in the query. This illustrates how embeddings capture the underlying meaning rather than merely relying on exact keywords. While the implementation of this process is beyond the scope of this article, you can learn more by checking out this YouTube video.
Fun fact - this example was generated by ChatGPT itself.
In essence, similarity searches operate by identifying the nearest numerical values of the search query to those of the source data. The closest match is returned. The process of transforming text into these numerical embeddings is also handled by the LLM.
Utilizing a vector store is most effective when handling a substantial amount of data. Given that six veterinarians can easily be processed in a single call to the LLM, I aimed to increase the number to 256. While even 256 may still be relatively small, it serves well for illustrating our process.
Veterinarians in this setup can have zero, one, or two specialties, mirroring the original examples from Spring Petclinic. To avoid the tedious task of creating all this mock data manually, I enlisted ChatGPT's assistance. It generated a union query that produces 250 veterinarians and assigns specialties to 80% of them:
-- Create a list of first names and last names
WITH first_names AS (
SELECT 'James' AS name UNION ALL
SELECT 'Mary' UNION ALL
SELECT 'John' UNION ALL
...
),
last_names AS (
SELECT 'Smith' AS name UNION ALL
SELECT 'Johnson' UNION ALL
SELECT 'Williams' UNION ALL
...
),
random_names AS (
SELECT
first_names.name AS first_name,
last_names.name AS last_name
FROM
first_names
CROSS JOIN
last_names
ORDER BY
RAND()
LIMIT 250
)
INSERT INTO vets (first_name, last_name)
SELECT first_name, last_name FROM random_names;
-- Add specialties for 80% of the vets
WITH vet_ids AS (
SELECT id
FROM vets
ORDER BY RAND()
LIMIT 200 -- 80% of 250
),
specialties AS (
SELECT id
FROM specialties
),
random_specialties AS (
SELECT
vet_ids.id AS vet_id,
specialties.id AS specialty_id
FROM
vet_ids
CROSS JOIN
specialties
ORDER BY
RAND()
LIMIT 300 -- 2 specialties per vet on average
)
INSERT INTO vet_specialties (vet_id, specialty_id)
SELECT
vet_id,
specialty_id
FROM (
SELECT
vet_id,
specialty_id,
ROW_NUMBER() OVER (PARTITION BY vet_id ORDER BY RAND()) AS rn
FROM
random_specialties
) tmp
WHERE
rn <= 2; -- Assign at most 2 specialties per vet
-- The remaining 20% of vets will have no specialties, so no need for additional insertion commands
To ensure that my data remains static and consistent across runs, I exported the relevant tables from the H2 database as hardcoded insert statements. These statements were then added to the data.sql file:
INSERT INTO vets VALUES (default, 'James', 'Carter');
INSERT INTO vets VALUES (default, 'Helen', 'Leary');
INSERT INTO vets VALUES (default, 'Linda', 'Douglas');
INSERT INTO vets VALUES (default, 'Rafael', 'Ortega');
INSERT INTO vets VALUES (default, 'Henry', 'Stevens');
INSERT INTO vets VALUES (default, 'Sharon', 'Jenkins');
INSERT INTO vets VALUES (default, 'Matthew', 'Alexander');
INSERT INTO vets VALUES (default, 'Alice', 'Anderson');
INSERT INTO vets VALUES (default, 'James', 'Rogers');
INSERT INTO vets VALUES (default, 'Lauren', 'Butler');
INSERT INTO vets VALUES (default, 'Cheryl', 'Rodriguez');
...
...
-- Total of 256 vets
-- First, let's make sure we have 5 specialties
INSERT INTO specialties (name) VALUES ('radiology');
INSERT INTO specialties (name) VALUES ('surgery');
INSERT INTO specialties (name) VALUES ('dentistry');
INSERT INTO specialties (name) VALUES ('cardiology');
INSERT INTO specialties (name) VALUES ('anesthesia');
INSERT INTO vet_specialties VALUES ('220', '2');
INSERT INTO vet_specialties VALUES ('131', '1');
INSERT INTO vet_specialties VALUES ('58', '3');
INSERT INTO vet_specialties VALUES ('43', '4');
INSERT INTO vet_specialties VALUES ('110', '3');
INSERT INTO vet_specialties VALUES ('63', '5');
INSERT INTO vet_specialties VALUES ('206', '4');
INSERT INTO vet_specialties VALUES ('29', '3');
INSERT INTO vet_specialties VALUES ('189', '3');
...
...
We have several options available for the vector store itself. Postgres with the pgVector extension is probably the most popular choice. Greenplum—a massively parallel Postgres database—also supports pgVector. The Spring AI reference documentation lists the currently supported vector stores.
For our simple use case, I opted to use the Spring AI-provided SimpleVectorStore. This class implements a vector store using a straightforward Java ConcurrentHashMap, which is more than sufficient for our small dataset of 256 vets. The configuration for this vector store, along with the chat memory implementation, is defined in the AIBeanConfiguration class annotated with @Configuration:
@Configuration
@Profile("openai")
public class AIBeanConfiguration {
@Bean
public ChatMemory chatMemory() {
return new InMemoryChatMemory();
}
@Bean
VectorStore vectorStore(EmbeddingModel embeddingModel) {
return new SimpleVectorStore(embeddingModel);
}
}
The vector store needs to embed the veterinarian data as soon as the application starts. To achieve this, I added a VectorStoreController bean, which includes an @EventListener that listens for the ApplicationStartedEvent. This method is automatically invoked by Spring as soon as the application is up and running, ensuring that the veterinarian data is embedded into the vector store at the appropriate time:
@EventListener
public void loadVetDataToVectorStoreOnStartup(ApplicationStartedEvent event) throws IOException {
// Fetches all Vet entites and creates a document per vet
Pageable pageable = PageRequest.of(0, Integer.MAX_VALUE);
Page<Vet> vetsPage = vetRepository.findAll(pageable);
Resource vetsAsJson = convertListToJsonResource(vetsPage.getContent());
DocumentReader reader = new JsonReader(vetsAsJson);
List<Document> documents = reader.get();
// add the documents to the vector store
this.vectorStore.add(documents);
if (vectorStore instanceof SimpleVectorStore) {
var file = File.createTempFile("vectorstore", ".json");
((SimpleVectorStore) this.vectorStore).save(file);
logger.info("vector store contents written to {}", file.getAbsolutePath());
}
logger.info("vector store loaded with {} documents", documents.size());
}
public Resource convertListToJsonResource(List<Vet> vets) {
ObjectMapper objectMapper = new ObjectMapper();
try {
// Convert List<Vet> to JSON string
String json = objectMapper.writeValueAsString(vets);
// Convert JSON string to byte array
byte[] jsonBytes = json.getBytes();
// Create a ByteArrayResource from the byte array
return new ByteArrayResource(jsonBytes);
}
catch (JsonProcessingException e) {
e.printStackTrace();
return null;
}
}
There’s a lot to unpack here, so let’s walk through the code:
Similar to listOwners, we begin by retrieving all vets from the database.
Spring AI embeds entities of type Document into the vector store. A Document represents the embedded numerical data alongside its original, human-readable text data. This dual representation allows our code to map correlations between the embedded vectors and the natural text.
To create these Document entities, we need to convert our Vet entities into a textual format. Spring AI provides two built-in readers for this purpose: JsonReader and TextReader. Since our Vet entities are structured data, it makes sense to represent them as JSON. To achieve this, we use the helper method convertListToJsonResource, which leverages the Jackson parser to convert the list of vets into an in-memory JSON resource.
Next, we call the add(documents) method on the vector store. This method is responsible for embedding the data by iterating over the list of documents (our vets in JSON format) and embedding each one while associating the original metadata with it.
Though not strictly required, we also generate a vectorstore.json file, which represents the state of our SimpleVectorStore database. This file allows us to observe how Spring AI interprets the stored data behind the scenes. Let’s take a look at the generated file to understand what Spring AI sees.
{
"dd919c71-06bb-4777-b974-120dfee8b9f9" : {
"embedding" : [ 0.013877872, 0.03598228, 0.008212427, 0.00917901, -0.036433823, 0.03253927, -0.018089917, -0.0030867155, -0.0017038669, -0.048145704, 0.008974405, 0.017624263, 0.017539598, -4.7888185E-4, 0.013842596, -0.0028221398, 0.033414137, -0.02847539, -0.0066955267, -0.021885695, -0.0072387885, 0.01673529, -0.007386951, 0.014661016, -0.015380662, 0.016184973, 0.00787377, -0.019881975, -0.0028785826, -0.023875304, 0.024778388, -0.02357898, -0.023748307, -0.043094076, -0.029322032, ... ],
"content" : "{id=31, firstName=Samantha, lastName=Walker, new=false, specialties=[{id=2, name=surgery, new=false}]}",
"id" : "dd919c71-06bb-4777-b974-120dfee8b9f9",
"metadata" : { },
"media" : [ ]
},
"4f9aabed-c15c-43f6-9dbc-46ed9a18e176" : {
"embedding" : [ 0.01051745, 0.032714732, 0.007800559, -0.0020621764, -0.03240663, 0.025530376, 0.0037602335, -0.0023702774, -0.004978633, -0.037364256, 0.0012831709, 0.032742742, 0.005430281, 0.00847278, -0.004285406, 0.01146276, 0.03036196, -0.029941821, 0.013220336, -0.03207052, -7.518716E-4, 0.016665466, -0.0052062077, 0.010678503, 0.0026591222, 0.0091940155, ... ],
"content" : "{id=195, firstName=Shirley, lastName=Martinez, new=false, specialties=[{id=1, name=radiology, new=false}, {id=2, name=surgery, new=false}]}",
"id" : "4f9aabed-c15c-43f6-9dbc-46ed9a18e176",
"metadata" : { },
"media" : [ ]
},
"55b13970-cd55-476b-b7c9-62337855ae0a" : {
"embedding" : [ -0.0031563698, 0.03546827, 0.018778138, -0.01324492, -0.020253662, 0.027756566, 0.007182742, -0.008637386, -0.0075725033, -0.025543278, 5.850768E-4, 0.02568248, 0.0140383635, -0.017330453, 0.003935892, ... ],
"content" : "{id=19, firstName=Jacqueline, lastName=Ross, new=false, specialties=[{id=4, name=cardiology, new=false}]}",
"id" : "55b13970-cd55-476b-b7c9-62337855ae0a",
"metadata" : { },
"media" : [ ]
},
...
...
...
Pretty cool! We have a Vet in JSON format alongside a set of numbers that, while they might not make much sense to us, are highly meaningful to the LLM. These numbers represent the embedded vector data, which the model uses to understand the relationships and semantics of the Vet entity in a way far beyond simple text matching.
If we were to run this embedding method on every application restart, it would lead to two significant drawbacks:
Long Startup Times: Each Vet JSON document would need to be re-embedded by making calls to the LLM again, delaying application readiness.
Increased Costs: Embedding 256 documents would send 256 requests to the LLM every time the app starts, leading to unnecessary usage of LLM credits.
Embeddings are better suited for ETL (Extract, Transform, Load) or streaming processes, which run independently of the main web application. These processes can handle embedding in the background without impacting user experience or causing unnecessary cost.
To keep things simple in the Spring Petclinic, I decided to load the pre-embedded vector store on startup. This approach provides instant loading and avoids any additional LLM costs. Here’s the addition to the method to achieve that:
@EventListener
public void loadVetDataToVectorStoreOnStartup(ApplicationStartedEvent event) throws IOException {
Resource resource = new ClassPathResource("vectorstore.json");
// Check if file exists
if (resource.exists()) {
// In order to save on AI credits, use a pre-embedded database that was saved
// to disk based on the current data in the h2 data.sql file
File file = resource.getFile();
((SimpleVectorStore) this.vectorStore).load(file);
logger.info("vector store loaded from existing vectorstore.json file in the classpath");
return;
}
// Rest of the method as before
...
...
}
The vectorstore.json file is located under src/main/resources, ensuring that the application will always load the pre-embedded vector store on startup, rather than re-embedding the data from scratch. If we ever need to regenerate the vector store, we can simply delete the existing vectorstore.json file and restart the application. Once the updated vector store is generated, we can place the new vectorstore.json file back into src/main/resources. This approach gives us flexibility while avoiding unnecessary re-embedding processes during regular restarts.
With our vector store ready, implementing the listVets function becomes straightforward. The function is defined as follows:
@Bean
@Description("List the veterinarians that the pet clinic has")
public Function<VetRequest, VetResponse> listVets(AIDataProvider petclinicAiProvider) {
return request -> {
try {
return petclinicAiProvider.getVets(request);
}
catch (JsonProcessingException e) {
e.printStackTrace();
return null;
}
};
}
record VetResponse(List<String> vet) {
};
record VetRequest(Vet vet) {
}
Here is the implementation in AIDataProvider:
public VetResponse getVets(VetRequest request) throws JsonProcessingException {
ObjectMapper objectMapper = new ObjectMapper();
String vetAsJson = objectMapper.writeValueAsString(request.vet());
SearchRequest sr = SearchRequest.from(SearchRequest.defaults()).withQuery(vetAsJson).withTopK(20);
if (request.vet() == null) {
// Provide a limit of 50 results when zero parameters are sent
sr = sr.withTopK(50);
}
List<Document> topMatches = this.vectorStore.similaritySearch(sr);
List<String> results = topMatches.stream().map(document -> document.getContent()).toList();
return new VetResponse(results);
}
Let's review what we've done here:
We start with a Vet entity in the request. Since the records in our vector store are represented as JSON, the first step is to convert the Vet entity into JSON as well.
Next, we create a SearchRequest, which is the parameter passed to the similaritySearch method of the vector store. The SearchRequest allows us to fine-tune the search based on our specific needs. In this case, we mostly use the defaults, except for the topK parameter, which determines how many results to return. By default, this is set to 4, but in our case, we increase it to 20. This lets us handle broader queries like “How many vets specialize in cardiology?”
If no filters are provided in the request (i.e., the Vet entity is empty), we increase the topK value to 50. This enables us to return up to 50 vets for queries like “list the vets in the clinic.” Of course, this won't be the entire list, as we want to avoid overwhelming the LLM with too much data. However, we should be fine because we carefully fine-tuned the system text to manage these cases:
When dealing with vets, if the user is unsure about the returned results,
explain that there may be additional data that was not returned.
Only if the user is asking about the total number of all vets,
answer that there are a lot and ask for some additional criteria.
For owners, pets or visits - answer the correct data.
The final step is to call the similaritySearch method. We then map the getContent() of each returned result, as this contains the actual Vet JSONs rather than the embedded data.
From here, it's business as usual. The LLM completes the function call, retrieves the results, and determines how best to display the data in the chat.
Let’s see it in action:
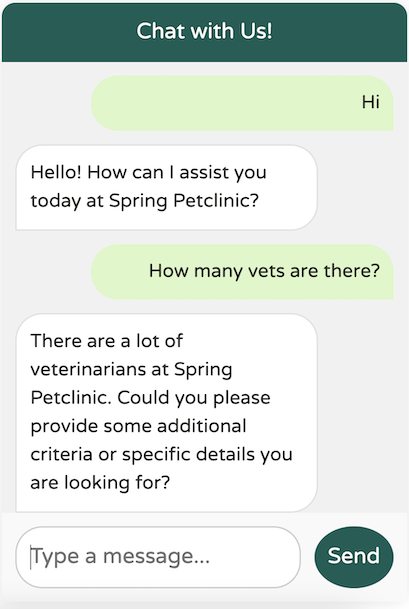
It looks like our system text is functioning as expected, preventing any overload. Now, let’s try providing some specific criteria:
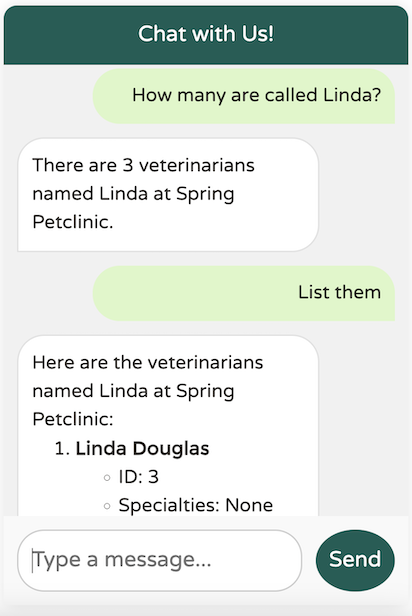
The data returned from the LLM is exactly what we expect. Let’s try a broader question:

The LLM successfully identified at least 20 vets specializing in cardiology, adhering to our defined upper limit of topK (20). However, if there’s any uncertainty about the results, the LLM notes that there may be additional vets available, as specified in our system text.
Implementing the chatbot UI involves working with Thymeleaf, JavaScript, CSS, and the SCSS preprocessor.
After reviewing the code, I decided to place the chatbot in a location accessible from any tab, making layout.html the ideal choice.
During discussions about the PR with Dr. Dave Syer, I realized that I shouldn't modify petclinic.css directly, as Spring Petclinic utilizes an SCSS preprocessor to generate the CSS file.
I'll admit—I’m primarily a backend Spring developer with a career focused on Spring, cloud architecture, Kubernetes, and Cloud Foundry. While I have some experience with Angular, I’m not an expert in frontend development. I could probably come up with something, but it likely wouldn’t look polished.
Fortunately, I had a great partner for pair programming—ChatGPT. If you're interested in how I developed the UI code, you can check out this ChatGPT session. It’s remarkable how much you can learn from collaborating with large language models on coding exercises. Just remember to thoroughly review the suggestions instead of blindly copy-pasting them.
After experimenting with Spring AI for a few months, I’ve come to deeply appreciate the thought and effort behind this project. Spring AI is truly unique because it allows developers to explore the world of AI without needing to train hundreds of team members in a new language like Python. More importantly, this experience highlights an even greater advantage: your AI code can coexist in the same codebase as your existing business logic. You can easily enhance a legacy codebase with AI capabilities using just a few additional classes. The ability to avoid rebuilding all your data from scratch in a new AI-specific application significantly boosts productivity. Even simple features like automatic code completion for your existing JPA entities in the IDE make a tremendous difference.
Spring AI has the potential to significantly enhance Spring-based applications by simplifying the integration of AI capabilities. It empowers developers to leverage machine learning models and AI-powered services without needing deep expertise in data science. By abstracting complex AI operations and embedding them directly into familiar Spring frameworks, developers can focus on rapidly building intelligent, data-driven features. This seamless fusion of AI and Spring fosters an environment where innovation is not hindered by technical barriers, creating new opportunities for developing smarter, more adaptive applications.